@outrigger
HA! You post a link from 1998!?
… and you think it even says what you think it says? Did you actually read it?
How about THIS article below! … which even if you are skeptical about the key placental link from South America to Australia, the article compares three very diverse “kinds” of Australia’s fauna and concludes that they are unusually closely related, genetically, despite the distinct “kinds” of animals they represent:
- a vegetarian mole;
- a non-jumping bandycoot type (omnivore); and
- a carnivorous hunter type (Tazmanian Devil).
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
This post is just to provide the separate link of the study mentioned above:
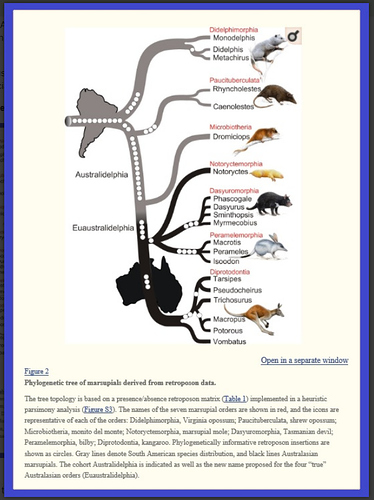
“Tracking Marsupial Evolution Using Archaic Genomic Retroposon Insertions” by Maria A. Nilsson, Gennady Churakov, Mirjam Sommer, Ngoc Van Tran, Anja Zemann, Jürgen Brosius, and Jürgen Schmitz
PLoS Biol. 2010 Jul; 8(7): e1000436. Published online 2010 Jul 27. PMCID: PMC2910653 PMID: 20668664
Notice in the image below, the various branches associated only with South America, and branches associated with Australia, where “radiating” speciation continued, in isolation from the rest of the world.
[Be sure to click on the images to enlarge text to a more convenient font size!]
.
.
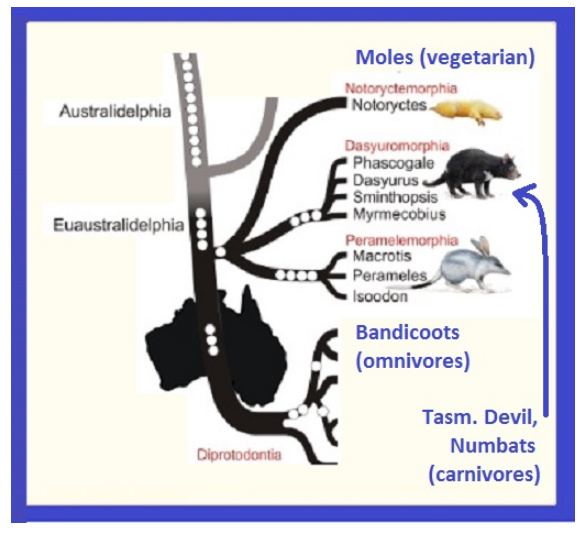
This image will be of particular value to us later on, because it creates a convenient grouping of some fairly disparate phenotypes:
While at the top we have “shrew-like” forms, and at the bottom we have “kanga” forms aggregated, in the middle grouping, we have the suggestion that three very distinct groupings share a close heritage:
Dasyuromorphia: the group having most of Australia’s carnivorous marsupials, including
quolls,
dunnarts,
the numbat,
the Tasmanian devil,
and the thylacine.
[In Australia, the exceptions include the marsupial moles and the omnivorous bandicoots.]
Notoryctemorphia: moles, vegetarian
Peremelamorphia: bandicoots & bilbies “the characteristic bandicoot shape: a plump, arch-backed body with a long, delicately tapering snout, very large upright ears, relatively long, thin legs, and a thin tail. Their size varies from about 140 grams up to 4 kilograms, but most species are about one kilogram, or the weight of a half-grown kitten [4 kilograms = 4 half-grown kittens].”
.
.
This is the ideal “research scenario” to see how much genetic change occurs, and how quickly - - according to Evolutionary Theory - - to accomplish divergence into three distinctive “forms” of marsupials!
.
.