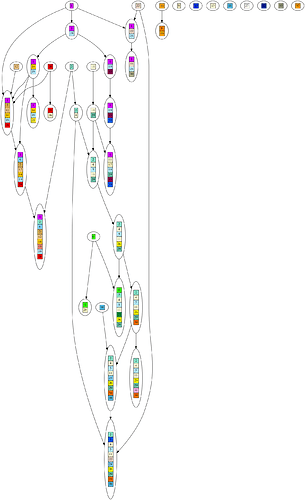
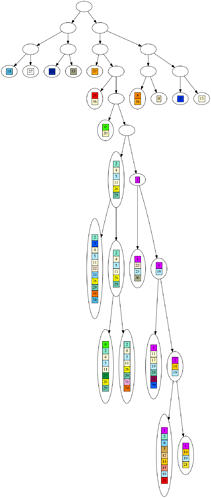
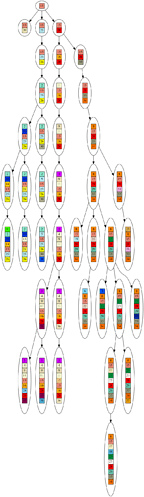
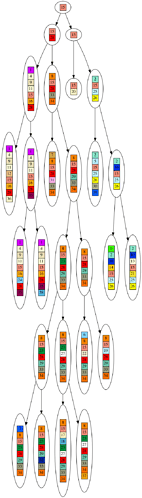
Here are some example graphs, so you know what the numbers refer to.
Example DAG:
Corresponding inferred tree:
Example evotree:
Corresponding inferred tree:
Example fake leaves (all same length, randomly sampled genes):
Corresponding inferred tree: