Thanks so much for your patient reply, Steve. I think it is starting to click now. I’ll have another read of your blog post and the related replies here and get back to you if I have any further questions (maybe by PM so as to keep this thread on track). Thanks again.
It is not just the GC content, but the frequences of each letter. For instance, the following string:
aaaatttggc
has frequencies:
a: 4/10
t: 3/10
g: 2/10
c:1/10
I compared a real human mtDNA to a real chimp mtDNA using the Levenshtein distance and got something that looked like your distribution, looking just at the replacement edits.
I then tabulated the atgc frequencies for the human mtDNA and used those frequencies to randomly generate another mtDNA of the same length. Same for chimp mtDNA. I then ran the same Levenshtein analysis and looked like I got a very similar distribution, except for the A<->C/G<->T bar.
I will be rerunning the analysis to make sure I didn’t make a mistake, and will post the code and graphs.
Side note, reason I went with Levenshtein first is because it was easiest way for me to reproduce what you did, but also the alignment algorithms have a substitution matrix which could bias the alignment to produce your distribution. I noticed in the hg19.panTro5.all.chain file that you linked had a substitution matrix that would favor alignments that would boost your transition bar:
A C G T
A 90 -330 -236 -356
C -330 100 -318 -236
G -236 -318 100 -330
T -356 -236 -330 90
Note the penalty is smaller for A-G and C-T alignments, which are the alignments that make up your transition bar. Since optimal alignment is computationally intractable for long DNA sequences, the algorithms have to use heuristics, and one heuristic is to assign penalties for different letter alignments. The above heuristic will assign smaller penalties for the alignments that make up your transition bar, so will possibly bias the inferred mutations towards your transition bar.
The alignment algorithms are similar to Levenshtein, but Levenshtein doesn’t make use of this heuristic matrix, so going with Levenshtein avoids this possible source of bias. I’m happy to see I still reproduced a distribution that looked similar to what you have, so the alignment algorithm didn’t introduce too much bias, if any.
Another side note, this use of heuristics in alignments, and then inferrence of evolutionary tree from said alignments, is what makes me somewhat skeptical about claims like ‘perfectly nested clades’. My understanding is these substitution matrices are essentially derived from existing human curated phylogenetic trees, and thus we have the possibility of data leakage into algorithmically generated trees from human constructed trees. Thus, the much acclaimed match between trees generated from genetic data and morphology could potentially be a case of what we call ‘data snooping’ in machine learning. In other words, the trees match because we’ve made them match. That’s why I’m interested in using metrics like plain Levenshtein distance where we don’t have the possibility of bias creeping into our results.
Bur for any decent-sized chunk of the genome (i.e. not the mtDNA), AT and GC are each split just about evenly between their constituent letters. The counts for chromosome 1 are:
T 67244164
A 67070277
C 48055043
G 48111528
So treating A=T and G=C is an excellent approximation.
Well, that’s not what I see in my mtDNA:
Counter(human)
Counter({‘c’: 5176, ‘a’: 5123, ‘t’: 4094, ‘g’: 2176})
Yes, as I said, mtDNA behaves quite differently. It has a very different replication system than the nuclear genome, with a meaningful difference between how the two strands are replicated and with substantial difference in base composition between them. In the nuclear genome, replication and transcription begin on both strands and there is no meaningful difference between the two ends of the chromosome or between the two strands – the content is different if you read in the opposite direction, of course, but the same machinery operates in both directions. As a result, any process that biases the base composition of one strand also operates on the other, and complementary bases occur equally frequently.
Can you unpack how this impacts the validity of my analysis?
I’m working on packaging everything up for independent verification, but here are my original numbers.
real human and chimp mtDNA comparison. not quite the same as in the article, but the same general trend we’d expect from biologically expected mutations of transitions > a/c&g/t > a/t > g/c :
len(transitions)
1206
len(a_t)
40
len(g_c)
19
len(a_clg_t)
69
Here are my numbers from the randomly generated sequences:
len(transitions)
1723
len(a_t)
1027
len(g_c)
722
len(a_clg_t)
1769
The numbers are not a perfect match, but there is the same general trend of transitions > a/t > g/c. The outlier is the a/c&g/t. So, at least part of the distribution in the article may be attributable to the gatc distribution. Another part might be due to the alignment heuristic matrix I mentioned previously. At any rate, it is far from clear to me the article distribution is a close shut case that differences between DNA sequences are explainable by biologically expected random mutations. My very sketchy analysis suggests there are a couple other possible explanations for the article distribution than biological mutations. I’d be very happy to be proven wrong, although I’d prefer it is in some way I can verify, vs just having to take someone’s word as authoritative.
As a side note, here is the weird thing I noticed comparing cross species in the a/c&g/t bar. Compare the following human/rat counts to the human/chimp counts above:
len(transitions)
1666
len(a_t)
638
len(g_c)
159
len(a_clg_t)
1253
We see the transitions > a/t > g/c. But whereas with human/chimp the a/c&g/t is much smaller than transitions, in this case they are very close. Yet, these are the mutations that are supposedly much less likely than the transition set. That’s why I made the claim there seems to be some sort of directedness here, since it at least notionally passes the explanatory filter: highly improbable differences that specify the species.
Also, I saw @T_aquaticus said my mention of detecting gmos as an example of design inference is goal shifting/bait and switch, not really the explanatory filter. Now, I know very little about GMO detection, but my general understanding is that there is a database of known GMO DNA sequences, and we basically look for pattern matches that are not attributable to chance, i.e. a BLAST search of a query sequence against the GMO database filtered by high bit scores. We can reframe this in terms of the explanatory filter:
- Eliminate chance/necessity with finding a highly improbable match
- Establish high specificity with an independent knowledge base, since the gmo sequences are human generated, not naturally evolved
Hopefully that makes it clear why I consider gmo detection a practical application of the explanatory filter that results in a design inference.
If you’re taking your single-base composition from mtDNA, you’re using the wrong values for anything other than mtDNA – which isn’t what you’re comparing your results to. In the human nuclear genome f(A) = f(T) = 29.5% and f(G) = f(C) = 20.5%.
Getting the order of 3 out of 4 numbers right, while getting their relative magnitudes quite wrong, is not a stunning success at modeling.
I showed you what the actual gatc distribution produces using your procedure. The main contribution in general of the composition to random comparisons should be to increase A<->T and decrease G<->C.
Quite unlikely, I think, given how very similar the human and chimp genomes are. There isn’t usually a lot of ambiguity about what the substitution is.
Before being proven wrong, you have to provide some reason to think that you might be right, which I don’t think you’ve done.
I don’t know what numbers you’re talking about here – are these your random comparisons, or what?
I’m referencing the real mtDNA, not the randomly generated sequences. If the distribution is due to mutation, then I’d assume it remains consistent between human/chimp and human/rat. Perhaps I’m missing something. One possible explanation is since human and rat are so evolutionarily distant, the common transitions are saturated.
Human/chimp:
len(transitions)
1723
len(a_clg_t)
69
Human/rat:
len(transitions)
1666
len(a_clg_t)
1253
I’ll have to recalibrate the numbers with your rate per base and see what that gives. Generally, the raw human/chimp Levenshtein comparison seemed to support the main feature of your first graphs, that the biologically expected mutations are the most common substitutions. In which case, it could be fair to say a lot of the genetic difference between human and chimps is due to random mutation.
In general, if I get persuaded that your distribution is a consistent feature in the genetic data we have across all species comparisons, I think that’s a pretty good piece of evidence in favor of random mutation explaining much of genetic difference. I’d at least have to come up with a good counter argument, so the ball is firmly in my court.
@glipsnort I believe I’ve persuaded myself of your argument, and addressed my counter argument.
When I normalize with base rates, I still get results a magnitude off of yours, although the transitions category remains the largest. That being said, it is a distribution reproducible across different species, we’ll see below:
$ python stats.py …/data/34-mammals/human …/data/34-mammals/chimpanzee
transitions 0.036430037426053365
a<->t 0.0021514629948364886
g<->c 0.0013067400275103163
a<->c|g<->t 0.0020825787758058673
However, now I can reproduce what you got with the random sequences, where we have a fairly equal distribution across the four categories:
$ python stats.py …/data/34-mammals/randgen_human …/data/34-mammals/randgen_chimp
transitions 0.05127972956658216
a<->t 0.06182271348071088
g<->c 0.046947397110929406
a<->c|g<->t 0.05779910660388748
And finally, across large species distances, like human/rat:
$ python stats.py …/data/34-mammals/human …/data/34-mammals/rat
transitions 0.050686056770817486
a<->t 0.03321186881832379
g<->c 0.0116406764770481
a<->c|g<->t 0.03781678785481761
I think in this case the transitions category gets saturated with mutations, and the other three categories maintain the same general distribution, although they become much closer to the transitions category.
Now, between more related species, we see the same sort of relationship between human/chimp, where transitions substitutions far exceed the other three:
$ python stats.py …/data/34-mammals/whiteRhinoceros …/data/34-mammals/indianRhinoceros
transitions 0.0461067704465108
a<->t 0.0059928086296444265
g<->c 0.0013199384028745324
a<->c|g<->t 0.0049909390689522
So, it looks like the mtDNA data backs up your argument that closely related species have the same substitution distributions, which is something we’d expect with random mutation, and not something we’d expect with guided mutation or genetic engineering.
For sake of addressing the last category of genetic engineering, here’s a chimera I created by patching the first third each of human, cat and finWhale mtDNA, and compared to human:
$ python code/stats.py data/34-mammals/human data/chimeras/human-cat-finWhale
transitions 0.033925686591276254
a<->t 0.03725216419994415
g<->c 0.035860655737704916
a<->c|g<->t 0.039310716208939146
Now we get the even distribution typical of random data, and it is distinctly different from the distribution we get comparing untampered sequences.
So, in summary, I’m pretty happy with your original article, since I can reproduce something akin to what you got, as well as address my own counter arguments. Armchair science!
Finally, for those who are interested in reproducing what I did, here is a repo complete with README.md, data and code: GitHub - yters/mutations: Compare mutation types between mtDNA.
Ah, got it. Yes, the mutation rate for mtDNA is high enough that saturation becomes a big issue when comparing between species. Even for human-chimp it must be noticeable.
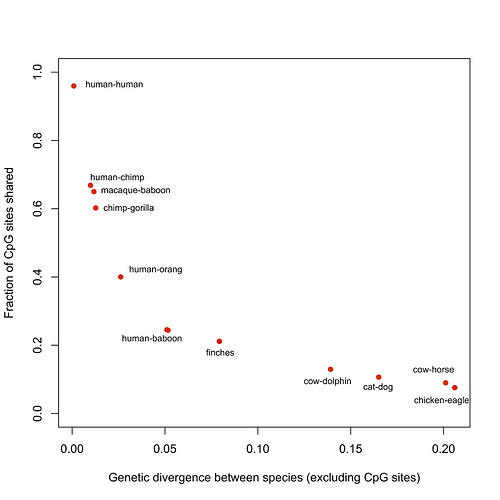
The same is true for nuclear CpG sites, which also have a high mutation rate – which is why I only showed the human-chimp comparison in my blog post. It’s easy to see how much more rapidly divergence increases as CpG sites than elsewhere:
Obviously, one can correct for saturation up to a point, but a complex analysis doesn’t make for an intuitively clear blog post.
Reality determines what effects a mutation will have. When there is evidence for mutations accumulating at a rate consistent with random fixation then we conclude those mutations are neutral. We follow the evidence.
Then how is it that neutral mutations become fixed in a population?
In 95% of the human genome, mutations accumulate at a rate consistent with random fixation. That’s the evidence.
Then show how the explanatory filter is used to detect GMO’s. Use that same method to compare the human and chimp genomes and show us which differences are due to design. As a control, compare mutations that we know have occurred in the lab or in the wild and see if the same method returns a negative for design.
All a BLAST search does is find the closest match between your sequence and the sequences in a database. That can’t be used to infer design.
More to the point, how did they determine which sequences go in the GMO database and which do not?
I would assume they know the specific patented sequences created by the GMO labs.
Also, the reason this is the explanatory filter and not just pattern matching is because BLAST provides an information score indicating how likely the match occurred by chance, i.e. eliminating the chance/necessity explanation.
This does not directly translate to design detection in the biological history record, since we don’t have a database of sequences used by gods or space aliens or whomever, but the point is it shows the explanatory filter works to detect design, which is the point being argued here.
This appears to be what you consider to be the explanatory filter. First, conclude something is designed. Second, put it in a database. Third, use a search function to see if something you have just found is in the database.
You don’t have a method for detecting design when you assume something is designed from the outset.
Assuming something is designed from the outset isn’t a good detector.
No, you are misunderstanding. The key thing that makes the process work is the GMO sequences are an independent knowledge source from evolutionary mechanisms. That’s why we can say they were designed instead of evolved.
Anyways, if you disagree, it would help me if you can point out how specifically the GMO example violates the explanatory filter. It seems to me to be exactly the explanatory filter resulting in a design inference.
How is the explanatory filter used to determine which sequences go into the GMO databases to begin with? Is the explanatory filter used at all?
I am also still waiting for an application of the explanatory filter to real world biological questions. We are told that some of the differences between the genomes of different species is due to design that happened millions of years ago. Where has the explanatory filter been used to determine this?
no that is not part of the explanatory filter
the important thing is the independence of the knowledge base, not the specifics of how it is constructed
and this is an example of a real world application of the explanatory filter to biology
Baloney. No one uses the explanatory filter to make the determination that the sequences in the GMO database are designed.