Despite the “hidden secrets” clickbait title, this seems like a pretty big deal, but I’d appreciate it if @glipsnort or @T_aquaticus (or anyone else with expertise) could explain Incomplete Lineage Sorting (ILS) in lay terms.
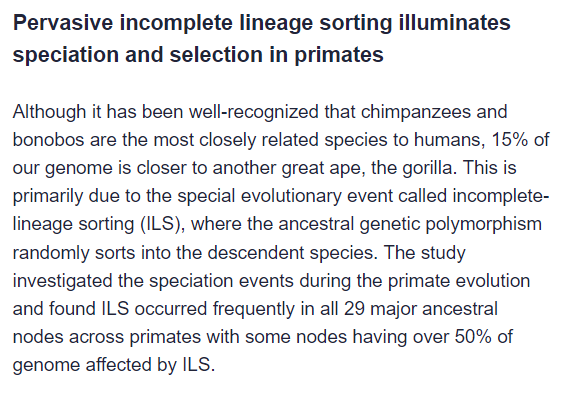
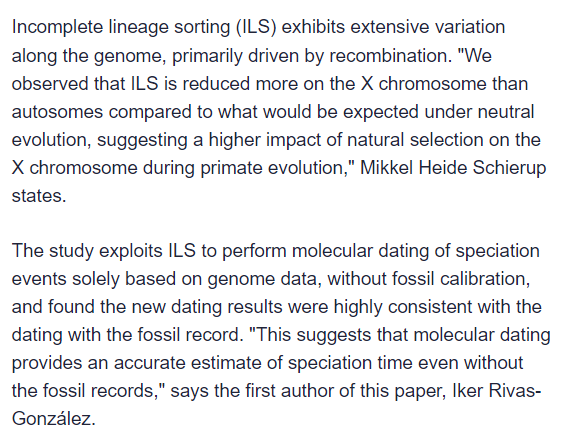
Despite the “hidden secrets” clickbait title, this seems like a pretty big deal, but I’d appreciate it if @glipsnort or @T_aquaticus (or anyone else with expertise) could explain Incomplete Lineage Sorting (ILS) in lay terms.


Isn’t this a counterexample to the claim of nested hierarchies? Ouch!
Once again the complexities of real life throw up a fog of confusion over our indisputable arguments. Though, it not as if the arguments were working anyway. LOL
I’m not sure if it’s the same as with plants. But I am under the impression what it means is that dna from an”family” level was found commonly in more than one genus and species but not the others. Or with primates, the ancestor of humans, chimps, bonobos and gorillas left parts of their dna that we may share with gorillas but we don’t share with chimps. But I am not 100% positive and look forward to what others have to say.
But I understand it also as sort of different subspecies of our primate ancestor that then intermingled with the main subspecies and then from there resulted in dna shared between various lower level clades.
Incomplete lineage sorting (ILS) is a pretty simple concept. I am assuming that most people learned Mendelian genetics at some point, and had to use Punnett squares:
In the case of ILS, there is a speciation event and the both new species are heterozygous for a given stretch of DNA, Gg from above. Another speciation event happens on one of the branches, and now we have 3 species that are all heterozygous, Gg. So something like this:
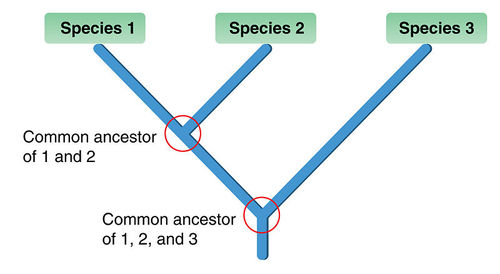
So all three species are still heterozygous, Gg. However, something interesting happens in species 2 after it splits off from species 1. It loses one of the alleles, and is now GG. Something also happens in species 1 and 3. In those species, the g allele fixes for gg. Now we have a case where species 1 and 3 share more DNA in one stretch of DNA even though species 1 and 2 are more closely related.
There’s also this article from back in the day on the main BL site:
https://biologos.org/series/evolution-basics/articles/incomplete-lineage-sorting
Specifically there, Venema notes:
The expected value of such (H,O) paired regions (~1.2%) is tiny when compared to the predicted value for (H,G) paired regions (around 25%), in large part because humans, chimpanzees and gorillas underwent speciation in a relatively short period of time, whereas the time between the orangutan divergence and the later gorilla divergence is greater. The genome-wide fraction of our genome that more closely matches the orangutan genome is about 0.8% – remarkably close to the predicted value, and consistent with the Ne values estimated for the (H,G,C) and (H,C) populations from prior work. In other words, when comparing primate genomes, we see a pattern of incomplete lineage sorting – as expected, our genome matches chimpanzees most frequently, then gorilla, and then orangutan.
By any chance do you know what it is called when you have something like
Plant A is the species.
It then has two subspecies such as Aa and Ab.
Then overtime Aa and Ab becomes its own distinct species but then one of them hybridizes with the original still existent A species so that it’s A x Ab and it’s seedlings are now AAb and overtime becomes its own main species again?
I was reading something a while back and can’t find it. It was more than just hybridization. It was like where a species got separated by a glacier and ended up developing into two subspecies then they lasted enough that the glacier was gone and the subspecies that’s now it’s own distinct species needs up hybridizing with the original species and overtime went through speciation and became its own species separate from the original and subspecies.
I believe that is called reticulate evolution.
From what little I have read about plant evolution, reticulation is relatively common in the evolutionary history of plants.
That’s it. I could not remember it for the life of me and typed in dozens of things. Thanks!
I was reading once where they thought that some of the chinquapins were actually a byproduct of reticulate evolution of American Chestnuts and others in the Castanea genus.
This is reticulate evolution and lost lineage sorting within the blackjack* pine. I always want to call the Pinus ponderosa the pond pine but that’s the P.serotina
I made it that far! Great explanation for those of us who sometimes struggle with simple concepts. haha
Don’t know why I didn’t think to search BL. Doh!
For the record, what I thought was a big deal in the OP was the project to “map” every primate’s genome. Who knows what that might reveal? Pretty cool.
If I understand @T_aquaticus correctly, I don’t think so. The other finding in the article above was that the molecular dating using ILS lined up with the fossil record.
I don’t trust Wiki or chatbots on subjects I don’t know very well. Why ask them when I have actual experts at my keyboard fingertips? Appreciate your time!
Exceptions are the rule in biology. Biology is messy, and this is one of the messy parts.
The scientific concept here is noise to signal. We know what can cause noise in phylogenies (i.e. deviations from the ideal signal). In this case, more closely related species will be impacted by ILS. As in the case of the great apes, ILS has a much greater impact on the trio of humans, chimps, and gorillas than it does on the trio of humans, chimps, and orangutans. There are actually pieces of the human genome that are more closely related to orangutans than they are chimps, but it is a relatively small amount, much smaller than the amount shared more closely with gorillas.
What we see is noise in within the group. What we don’t see is massive violations between very distant branches. For example, we don’t find exact copies of mouse genes in the human genome that are wildly different than the homologous gene in chimps and gorillas. The noise is where we would expect it to be, at least from my limited understanding of phylogenetics.
So in some sense we can say the exception proves the rule.
Good stuff. I’m curious what the data will show when every living primate’s genome is mapped and put through this process. It won’t necessarily affect our view of human evolution, but it could reveal a lot about the evolution of our more distant cousins.
In some sense, yes. With the caveat of “to the best of my limited knowledge” . . . if speciation events are close to one another then ILS will be more pronounced. For example, a snippet from the paper referenced at the beginning of the thread:
Gibbon species evolved close together in time, and the amount of ILS demonstrates this. The decreasing amount of ILS as you move further down in the great ape tree is also what we would expect, that is less human DNA closer to orangutans than to gorillas. So it’s not useless noise. The noise can actually tell us something about the evolutionary history of the group.
Paralysis by analysis is the first thing that comes to mind. ![]()
I need to go back and read over some of the helpful, explanitory comments. But this jumped off the page at me:
The study exploits ILS to perform molecular dating of speciation events solely based on genome data, without fossil calibration, and found the new dating results were highly consistent with the dating with the fossil record. “This suggests that molecular dating provides an accurate estimate of speciation time even without the fossil records,” says the first author of this paper, Iker Rivas-González.
Do I understand correctly that this means there are two, independent ways of dating speciation events, which consistently confirm each other?
If so, this should lead to some “interesting disputes.”
“Let your conversation be always full of grace, seasoned with salt, so that you may know how to answer everyone.” -Colossians 4:6
This is a place for gracious dialogue about science and faith. Please read our FAQ/Guidelines before posting.