An astonishing similarity of protein coding genes in humans, pigs, mice, dolphins, kangaroos, spiders etc.
https://proteomics.cancer.gov/whatisproteomics
Excerpt: “The human genome contains about 21,000 protein-encoding genes, but the total number of proteins in human cells is estimated to be between 250,000 to one MILLION.”
My comment: Newest studies have confirmed the number of human protein coding genes to be about 19,000. RNA-directed cellular mechanisms are able to build thousands of different proteins by using raw data of one gene, without changing the gene’s sequence. Mechanism is called alternative mRNA splicing. Based on DSCAM gene in Drosophila Melanogaster, the fruit fly’s cell is able to build even 38,016 different proteins, without changing the sequence. The way of how the cell uses protein coding genes tells us that genes are not drivers. Instead, they are just raw data, libraries for RNA-directed mechanisms.
The alternative splicing is the most significant mechanism inducing the ecological adaptation and organisms’ variation. The clever question arises: how come variation of organisms happen if the sequences of protein coding genes are not to be changed? Mutations in protein coding genes could be extremely harmful and they would cause a huge mess in genomic stability and integrity.
Serious science has the answer:
Epigenetic Control of Gene Expression. The alternative splicing mechanism is regulated by several epigenetic factors. The three main regulators are:
-
The DNA methylation.
The alternative role of DNA methylation in splicing regulation: Trends in Genetics -
MicroRNA downregulation.
https://www.lakeforest.edu/live/news/4451-co-regulation-of-mirna-biogenesis-and-alternative -
Histone methylation
Histone methylation, alternative splicing and neuronal differentiation - PubMed
By the way, did you know that the C. Elegans, a tiny multicellular nematode worm, that is assumed to be a simple type of organism, has 20,450 protein coding genes? More than us! It is not a simple life form! It also uses the alternative splicing machinery for regulating its proteins in 1031 cells it has.
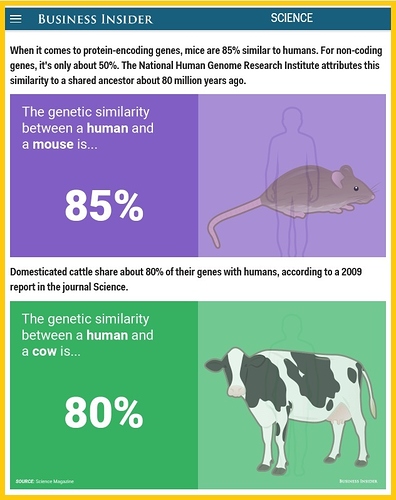
Protein coding genes are very similar in most animals. For example, human and mice genomes differ only at 2.5%.
Human and pig genomes are also extremely similar.
http://www.nationalhogfarmer.com/news/human-to-pig
“We took the human genome, cut it into 173 puzzle pieces and rearranged it to make a pig,” explains animal geneticist Lawrence Schook. “Everything matches up perfectly. The pig is genetically very close to humans.”
Human protein coding genes are also very similar to kangaroos, dolphins, spiders etc. These are very inconvenient facts for believers of the evolutionary theory that is misleading people by maintaining the heresy of human-chimp genomic similarity. They don’t tell you that the famous 98% is true only with these protein coding genes. But when we compare the alternative splicing mechanism and epigenomes, the differences are quite remarkable.
Everything points to Design and Creation. Don’t get misled.