@jpm
So, let’s imagine doing this kind of analysis, starting with all the animals we can look at today, right now.
If we start ticking the Genetic Clocks back into history, we are looking for when 2 (or 3 or a dozen) cousin “branches” of existing species merge into a common ancestral population.
What this is telling me is that 100,000 to 200,000 years ago, very broad ecological shifts, and even shifts in geographical territories, were “hitting” existing populations hard … and thus creating the necessary changes in selection pressures needed to “expose” the sudden value of other adaptations!
150,000 years ago, all the existing populations had their baggage of variant alleles - - in a genetic “come as you are” party! Some populations had a lot of them, and some didn’t have as many. We know that every time a population becomes environmentally stressed, it exposes the hidden value of all sorts of configurations of alleles - - and configurations of alleles - - that never really mattered before.
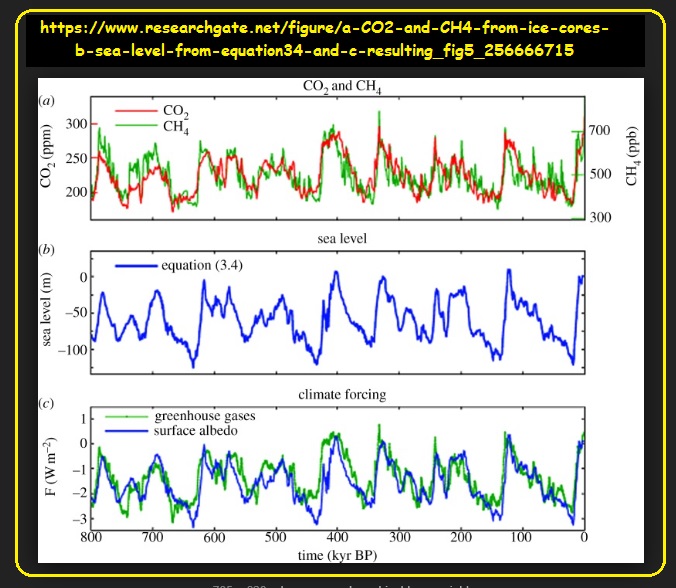
So… what might do that? Well, for 800,000 years, Earth has been globally subjected to Ice Ages - - pretty much one every 100,000 years!
This makes good sense, yes?
Only surviving populations are available to analyze. So I can only surmise that each one of these Ice Ages created its own batch of “new surviving” populations.
And from these new populations created anywhere from 800,000 to 300,000 years ago, the most recent ice ages (100,000 or 200,000 years ago) produced the “spanking new” populations with genes we can analyze!