In previous posts I discussed the position of ERVs in genomes, and how they evidence common ancestry between humans and other apes. Here is an excerpt from a 1999 peer reviewed paper by Johnson and Coffin:
“Given the size of vertebrate genomes (>1 × 109 bp) and the random nature of retroviral integration (22, 23), multiple integrations (and subsequent fixation) of ERV loci at precisely the same location are highly unlikely (24). Therefore, an ERV locus shared by two or more species is descended from a single integration event and is proof that the species share a common ancestor into whose germ line the original integration took place (14).”
http://www.pnas.org/content/96/18/10254.full
As with all good science, it is always a good idea to test a theory from multiple independent lines of evidence. ERVs actually supply 3 different lines of independent evidence for common ancestry, one of which I discussed in previous posts. The next line of independent evidence is LTR divergence. From Johnson and Coffin (1999):
“Third, sequence divergence between the LTRs at the ends of a given provirus provides an important and unique source of phylogenetic information. The LTRs are created during reverse transcription to regenerate cis-acting elements required for integration and transcription. Because of the mechanism of reverse transcription, the two LTRs must be identical at the time of integration, even if they differed in the precursor provirus (Fig. 1A). Over time, they will diverge in sequence because of substitutions, insertions, and deletions acquired during cellular DNA replication.”
Here is a diagram of a retroviral genome:
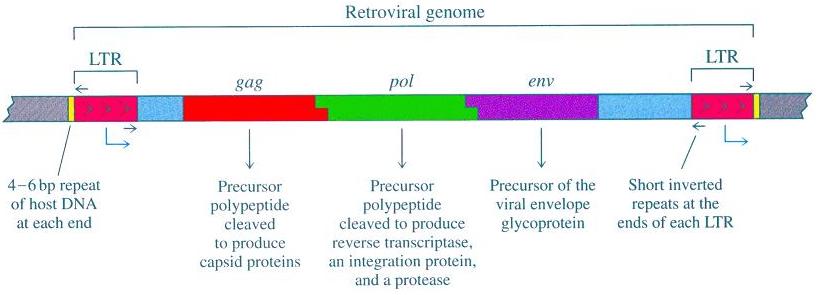
As you can see, the LTRs, or long terminal repeats, are like the bookends of the retroviral genome, called the 5’ and 3’ LTRs. As part of the retroviral life cycle, one LTR on one side is copied to produce the other LTR on the other side. Therefore, when the retrovirus inserts into the host genome the two bookends (the LTRs) have identical sequence. Once in the host genome, the slow accumulation of different mutations in each LTR will cause the LTRs to become less similar over time.
So how does this phenomenon allow us to test common ancestry? Evolution predicts that LTR sequences should reflect evolutionary distance, and that they should diverge from each other. That is, there should be more differences between the orangutan and human LTRs than between the chimp and human LTRS. If we use LTR sequences as inputs into phylogenetic algorithms then the phylogenies produced by these algorithms should match the phylogenies constructed by morphology (i.e. what the species look like). This is the standard phylogeny for apes:

From the Johnson and Coffin (1999) paper, this is the phylogeny of shared LTRs among different primate species:

(click on image for larger version)
As you can see, the standard phylogeny and the LTR phylogenies match for almost all nodes. On top of that, the 5’ and 3’ LTRs are on different branches of the phylogeny, consistent with the expected divergence. This is yet another independent line of evidence that screams common descent. Only evolution predicts a phylogeny of LTR sequences. ID/creationism does not.