Just to give another example of what biologists are seeing.
Most genes in vertebrate genomes have introns. These are long stretches of DNA that are clipped out of the RNA transcript from that gene. The exons in a gene contain the coding regions that are ultimately translated into protein.
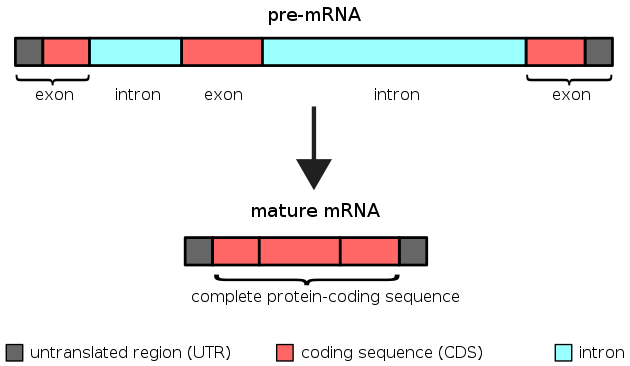
The vast majority of DNA sequence within introns doesn’t affect its function. The sequence immediately surrounding a splice site matters, and there are a few functional sequences found in introns here and there, but for the vast majority of intron sequence the sequence can be entirely random and still function as an intron since it is spliced out and doesn’t affect the resulting coding sequence.
Knowing this, what would we expect to see if common ancestry and evolution is true? First, we would expect mutations to happen in both exons and introns. However, mutations that occur in exons will have a much, much higher chance of being deleterious and will therefore be selected against by natural selection. Since the DNA sequence doesn’t matter for the vast majority of intron sequence, very few deleterious mutations will occur in intron sequence. This means we would expect exons to accumulate fewer mutations over time than introns. This means we would expect to see more differences in intron sequences when we compare the same gene between two distantly related species. We would also expect this difference to increase with evolutionary distance.
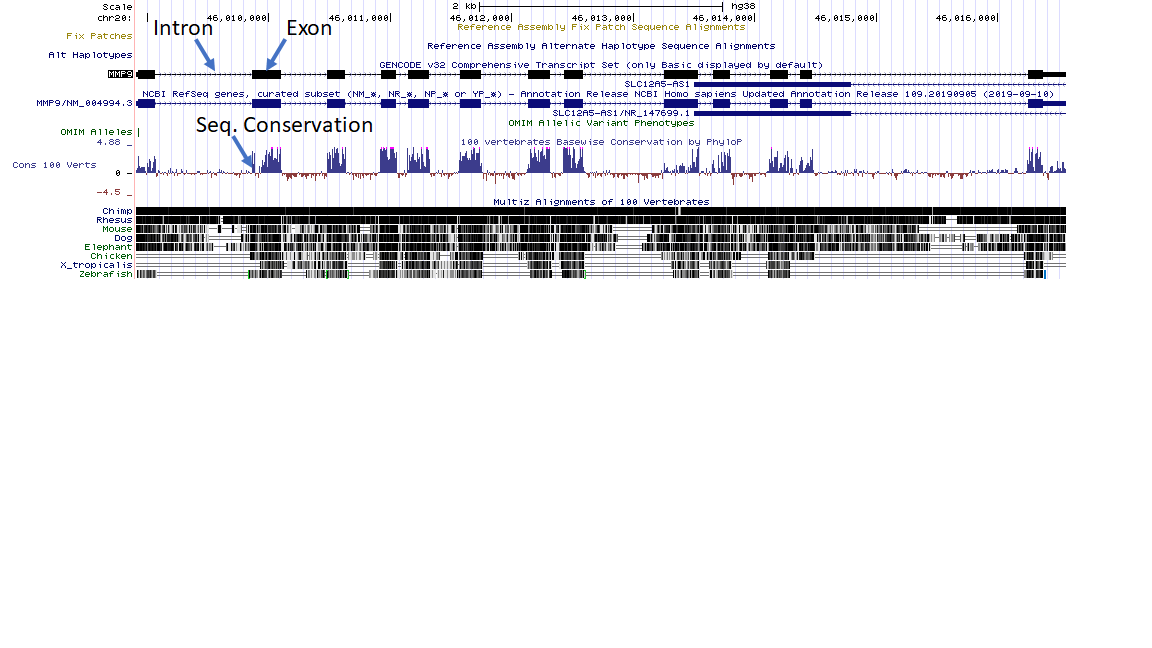
Guess what? This is exactly what we see (click for larger version).
This was downloaded from the UCSC genome browser, and the gene we are looking at is human mmp9, a gene shared by a wide range of vertebrates. The boxes are the exons, and the line connecting the boxes are the introns.
Below the gene is a graph showing sequence conservation across 100 different vertebrate genomes. The higher the line goes the more sequence conservation there is. Notice how the graph spikes where there is exon sequence, and flatlines where there is intron sequence.
There are also a few model vertebrate species listed below. Where there is black there is matching sequence. Notice how the entire bar is black across the chimp genome. Again, a close relative of humans has sequence very similar to humans. However, as you increase evolutionary distance you will notice that there are fewer and fewer similarities in the intron sequence while there is still noticeable sequence similarity in the exons. When you get to the zebrafish and frog (Xenopus tropicalis) the only sequence similarity that can be consistently found is in the exons.
This is EXACTLY what we would expect from evolutionary mechanisms. I have yet to see anyone explain this data outside of evolution. For example, if species were separately created then why not use identical introns for fish and humans since the sequence really doesn’t matter? Why should introns differ more than exons if life did not evolve and does not share common ancestry?
note: Inspiration for this post comes from “Faith and the Human Genome” by Dr. Francis Collins. He discusses the same patterns for a different gene.