This is just so obvious that it is worth including the figures here. With the relevant caption…
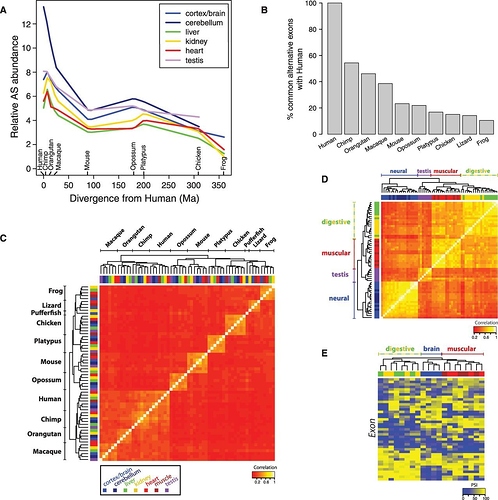
Profiling of alternative splicing (AS) in vertebrates. (A) Relative proportions of exons undergoing AS in each sample, … (y-axis units relative to the sample with lowest AS frequency). … (B) Percentage of common AS events between human and other species., (C) Symmetrical heat map of Spearman correlations from PSI [splicing] profiles. For each sample, PSI values for the 1550 orthologous exons in the 11 analyzed species were estimated.
Reading this you can see figure 1A shows approximately equal amounts of alternative splicing for all samples except human cerebellum. Other than cerebellum, all other tissues measured have LOWER splicing than chimps.
Reading figure 1B, it is important to understand the axis first. Notice that humans have 100%? That is not because 100% of exons are alternatively spliced, but because 100% of human alternative spliced exons are alternatively spliced in human (by definition). The rest of the bars just show how many exons are spliced in both humans and each of the other species. About 50% are shared with chimps. The rest are not.
But we already know that there is a very large number of changes in the cerebellum (from Fig 1A). Looking at the high correlation (of about 50%) in figure 1C, we can be certain the bulk of the differences are in the cerebellum. Of course the data is available, so you can go test it yourself.
To be clear, this data is particularly problematic for any model that does not accept common descent.
First, there are a large number of “skipped” exons in humans and other species. We still see the exon, but is not used. Why is it still there? It is a lot like a pseudogene. Let me coin a term: a pseudoexon. This makes a ton of sense in common descent.
Second, why is that alternative splicing patterns are so species specific (Fig 1C)? In contrast, as a control, gene expression is much more tissue specific (Fig 1D)? This is exactly the same pattern (species specific rather than function specific) we see in neutral mutations (e.g. synonymous mutation) too. It is a clear signature of common ancestry.
Of course, we know that you (@Cornelius_Hunter) do not appreciate this evidence. That is fine. But what is your alternative explanation? Why do you see the patterns here that you do?
We keep asking for your model. We never get it…