Good point. I’ll try to do better.
Did you forget Post #13 where you said:
The point that I would like to make for your consideration is that for each small random change in the DNA, a corresponding change must also occur in all the other sub-systems that depend on the one that experienced the random change if they are to remain functional at the higher “system of systems” level. You simply cannot change a single protein even if it only results in a minor fold change or makes it unstable so that multiple folds are possible. The effect of the change cannot be realized unless all the other proteins change in very specific ways that enable the required coordination to continue in the cell that depend on that protein’s function in all of its states.
This was completely wrong and the scenario was from you superimposing your SE background onto Biology. i.e. you made up a scenario that doesn’t actually exist in nature and used it as strong evidence that modern biologists are rather foolish.
I see that it’s that you guys don’t agree with me rather that it is wrong. You and others seem to have missed the key words in my claim. Those word are: “…in all the other sub-systems that depend on the one that experienced the random change…” If one sub-system doesn’t depend on the one that changed, then you are correct. But I was very specific in saying that it did depend on it. If it’s unaffected by the change, then it’s not dependent on it. Simple logic. Of course, one sub-system dependent on another may only be so for a restricted range of inputs, but in general, if it’s dependent, it’s affected by the change. Where am I going wrong?
Raymond, not getting your facts straight is not a matter of agreement or disagreement. It is a matter of not getting your facts straight.
Or if I told you that one plus one equals three, and you told me that I was wrong, it’s two, would that be just you disagreeing with me?
Raymond, you’re going wrong in the points that you’re not addressing as much as in the points that you are. I’ve mentioned the concept of canary deployments two or three times in this thread – a concept that completely changes the dynamics of how sub-systems affect each other, and also completely changes how systems engineering relates to the subject of evolution. Yet you haven’t addressed them once. In fact, you haven’t even demonstrated that you understand the concept at all.
Not to be too picky about it, but you quoted Philip Johnson who quote mined Gould to make a point that Gould wouldn’t agree with. Any author that is guilty of quote mining, in particular a quote mine that is done so often it is on the list, is not to be trusted. At least in my opinion.
In your world of requirements you would be correct. In the real world he was indicating the expected results. And if you have done any reading of Gould you would know that is what he would expect.
This is Quote #14 on the Quote Mine Project list.
Raymond if you are not aware, The Panda’s Thumb is written for a general audience. It is not a technical paper or book.
Hi Raymond,
I want to start by owning up to a mistake that I made. Given the following assumptions:
- peptide bond probability is relevant
- chirality is relevant
- Proteins shorter than 150 amino acids are not viable in a protobiotic environment
- Rumraket’s 10e-3 estimation of the density of functionality in proteins is correct
The probability formula is:
10e-3 * 2e-150 * 2e-150 ~= 10e-93
We can also substitute Hunt’s and Axe’s estimations for the first term and arrive at probability estimations of 10e-100 and 10e-167 respectively.
I note that we are in agreement about Axe-based and Rumraket-based estimations now, given identical assumptions.
By owning up to my mistake, I trust that I have dispelled any notion of being motivated by “personal incredulity” or clinging to an evolutionary narrative in advancing the arguments I made.
Also, I want to mention that I do not watch YouTube videos for several reasons:
- Inefficiency of information transmission:
- I read far more quickly than I can watch a video.
- When reading, I can skim the parts I am already familiar with or are not relevant to the topic I am researching. This is vastly more difficult with YouTube videos.
- Quality control: It’s hard to find counter-arguments to any particular YouTube presentation by searching on the presentation/title, because most specific counter-arguments–to the extent they exist–are also YouTube videos. And the text of YouTube videos does not (yet) show up in Internet search indexes.
- Suspension of critical facilities: When I read, I am able to step back, ask questions, and reflect on what I’m reading. When I watch a video, my attention is fully occupied by the video presentation, leaving me no mental capacity for reflection until after the video has finished.
So I don’t watch videos. But I have read pretty much all your links to articles, Raymond.
Your truncation of my argument omits extremely important context. Allow me to quote the relevant portions of the argument:
The subject I was discussing was the inference to theory that scientists in several fields (geology, astronomy, astrophysics, biology) use under the condition of incomplete understanding of mechanisms.
Your reply did not deal with the subject of inference to a theory of origins under the condition of incomplete understanding of mechanisms. Instead, you chose to talk about the value of SE in understanding cellular mechanics. If you cannot see why this is a different topic, I’m not sure how we can make progress.
Raymond, I quoted your original statement in which you complained about the use of extrapolation in biology. Curiously, you chose to omit the fact that I was relying on your quote. How can I be creating a straw man when I was quoting you?
If you would like to clarify how it is that, for example, you think biology uses extrapolation differently in the theory of evolution than astrophysics uses extrapolation in the theories of stellar formation and galactic formation, then we can have a reasonable discussion. But you were using “extrapolation” as a code word for “unscientific.” That’s why I said what I said.
Sure. I did read the ENV article by Jonathan Wells, which struck me as an extended argument from incredulity. (So many changes! How could it happen?) Consider, though: There is a coordinating force at work. The aquatic environment, which promises an abundant food source to mammals that can explore it, exerts selective pressure on the phenotypes of populations of mammals that are exploring it. And that selective pressure indirectly can produce coordinated genotype changes.
Allow me to illustrate with an example in human evolution among the Bajau people of the South Pacific. Coastal-dwelling Bajau are able to stay underwater far longer and dive to far deeper depths (200 feet!) than nearby populations. Dr. Melissa Ilardo has identified specific mutations in the PDE10A gene and the BDKRB2 gene which have led to two specific adaptations among the Bajau: 50% larger spleens, and constriction of superficial blood vessels while diving. It’s a fascinating study:
And these changes occurred in a period of just 500 - 10k years.
The Bajau data are only suggestive, however. Do we have anything more specific on the whales? It turns out that very recent research has made considerable progress in identifying the genetic and molecular changes involved in cetacean evolution. (Note: all articles I cite are freely available on the web.)
In conclusion, a total of 17 residues were replaced on or near the protein surface of Mb, during the evolution from the terrestrial ancestor to deep-diving extant whales through the intermediate ancestor. The time range of the early phase of evolution from the terrestrial (aMbWp) to the intermediate (aMbWb or aMbWb’) is assumed to be ~10 M years, from the early Eocene to the middle Eocene, and that of the late phase from the intermediate to the sperm whale (swMb) is estimated to be approximately 40 M years from the middle Eocene to the present. Thus, the Z Mb and the precipitant tolerance (β) had evolved first and rapidly by the nine residue replacements, whereas the repulsive interaction had evolved subsequently and rather slowly by the ten residue replacements. The correlation between Z Mb and β is evident, but still it is not clear whether the Z Mb increase alone could sufficiently explain the β increase or not. The physicochemical mechanism for the β enhancement is an important subject to be tackled in the future.
Four lines of evidence support the hypothesis that significantly adaptive molecular evolution of cetacean osmoregulation has occurred. First, the selection analysis showed that four genes involved with osmoregulation, i.e., ACE, AGT, AQP2, and SLC14A2, were subjected to strong positive selection in cetacean-specific lineages, whereas no selection was observed in terrestrial mammalian lineages such as cetartiodactyls, carnivores, rodents, and primates. Second, adaptive evolution was further supported by evidence that radical changes in amino acids have occurred significantly more often than conservative changes in cetacean osmoregulation-related genes. Third, the positively selected sites were localized in, or close to, the functional regions in the predicted 3D structures of the ACE and SLC14A2 genes. Finally, 13 codons (ACE: 246, 315, 808, and 836; AGT: 260; SLC14A2: 27, 54, 80,169, 395, 397, 672, and 728) exhibited parallel amino acid changes in different cetacean lineages (Figure 1). Overall, the significantly positive selection identified in cetaceans suggests an enhancement of their osmoregulation capacity when they shifted their habitat from the land to the sea.
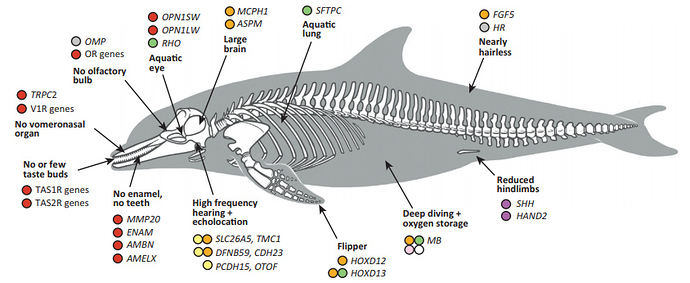
And here is an impressive mapping of the genetic and phenotypic adaptations in dolphins
And here is a genomic analysis of nested hierarchy within the dolphin clade of cetacea:
https://www.pnas.org/content/98/13/7384
It’s an interesting experience to be told this by a program manager who exhibits fundamental misunderstandings of microservice architecture and no apparent knowledge of or even interest in the mathematical foundations of deep learning algorithms. Yet both of these system architectures provide key insights into biological systems.
The thing about systems is that different kinds operate under different sets of constraints and objectives. It is not necessarily the case that principles in mechanics, fluids, gases, and material science are more relevant to biological systems than principles of microservices or the mathematics of neural networks. You can’t simply assume that one set of systems is more relevant than the other. You have to show that the features of biological systems that have been enumerated by biologists are more proximate to, say, mechanics than to local search algorithms.
And here you have paved the off-ramp for my participation in the discussion. When someone hurls insults and taunts, it’s difficult to see how they can be open to reasoned discourse. It would be fine to say I misunderstand something or even that I have fundamental misunderstandings of something. But puerile is a slam, an insult, a pejorative that has no place in reasoned discourse–especially among Christians.
Have a blessed week, my brother –
Chris
EDIT: Here are some synonyms for “puerile” from thesaurus.com:
- infantile
- babyish
- naive
- foolish
- ridiculous
- silly
- weak
EDIT 2:
Your repeated use of “sixth grade show and tell” to describe what you disagree with is also highly pejorative, and a strong signal of likely being in a poor frame of mind for reasoned discourse.
Are you (at least implicitly?) agreeing that this evidence is highly congruent with common ancestry?
But aside from that, I’m simply asking what YOUR explanation is for these data. Why (in your mind) do we see this over and over again in nature?
You were introducing the use and acceptance of extrapolation in other scientific disciplines as though to authenticate its use to support evolution in biology. That’s a classic strawman. Extrapolation is only valid where the specific problem and data warrant it.
That strikes me as heavily biased.
Do these coordinating forces lesson the randomness of the foundation of evolution, i.e., random change?
How does this show that evolution is a better explanation? How do we rule out design? Can God have included a epigenetic mechanism in His design to ensure the divers could adapt to this environment to survive? I’m not sure why you offered all the details of this? How does that rule out design?
Ditto on the Cetacean example. How does this rule out design?
I still don’t understand how you could conclude that I fundamentally misunderstand microservices and deep learning algorithms. I told you I did, and my last two years before retirement as a Research Scientist at NGA I focused on deep learning neural nets to solve a set of challenging problems we had in image exploitation. My leadership even directed me to take it to DARPA. Do you think they would do that if I didn’t understand it? By the way, should I not take this as insulting? For sure it’s uninformed, but who uses ignorance as the basis to insult someone? There are some colorful words to describe them, but since you have a thin skin, I’ll leave it at that.
That may be for the best since most of what you have offered hasn’t been constructive. You seem to be more focused on winning the argument for evolution than digging into uncover the important issues. You have attacked me more than anyone (ad hominem), and mostly based on being uninformed. You insist that your “data science” view of the world trumps all others, especially the “physical science” view. Clever algorithms reflecting recent advances in computer science seem to color your judgment about everything else, and those who are not in your circle are lessor beings. I have endured your subtle insults, and will continue since my goal is to understand evolution to see if indeed it is a better explanation.
Yes, it appears consistent with the HYPOTHESIS of common ancestry. Questions remain: "When we have data that is “consistent with” a hypothesis, does that make it conclusive? If not, what work remains to make it conclusive? How does this show that evolution is a better explanation than design?"
I’m trying to draft some thoughts on how we can answer these questions, and I look forward to engaging with you on it. Maybe I can get those thoughts out later today.
Hi Raymond,
I provided links to five freely available articles, and only one of them got clicked before your reply.
I also note that you did not take the opportunity to own up to your use of taunts and insults.
I do pray for God’s blessings on you as we go our separate ways.
Chris
I’m looking for evidence that’s not just anecdotal, but substantive that can be used to confirm, not just “be consistent with.” When you can provide something that shows clearly macro-evolution is occurring without having to be imaginative, I’ll read it. Give me something to read that will clearly distinguish between something that results from “mutation/natural selection” and something that is the result of epigenetic adaptation. How would you or anyone else (since you’re leaving) decide that?
Hi Ray,
All of the papers and articles deal with changes in nucleotide sequences. None of them deal with changes in expression due to methylation.
Chris
Ceteris Paribus?
No methylation at all.
It’s good to see that even after a conversation is obviously dead, there can still be talk about methylation. It’s truly transgenerational! 
A post was split to a new topic: How are bacteria flagella duplicated in reproduction?
They have done the same thing when Darwin Devolves and the science review
NOTHING in that post supports any of those claims. Also, Wells butchers paleontology. I have seriously cried from laughing so hard at some things he has said. If you want to talk whales come to someone who has worked or works on them (me. I have literally held some of the fossils in my hand)
A lot of cetacean evolution happened through the formation of pseudogenes:
https://www.sciencedirect.com/science/article/pii/S0169534714000846
Thanks. I’ll look at the reference, and get back to you. I look forward to digging into this as it seems to be an important issue with regard to our discussion on evidence.
If you have any particular notes on the Wells article especially where you think he’s wrong, I would find that helpful as examine both sides. I give them as hard a time as you guys. I’m learning a lot, and I really appreciate the contributions of all of you.
I think I could have used the Latin phrase “quid erat demonstrandum” after post #414. On further reflection, I am not even sure what “ceteris” you had in mind that would be “paribus.”