I’m going to add your name to the next edition of this book
hi @dcscccc
Hope your Easter was full of joy. My wife and I traveled to visit our daughter over the weekend, so I was unable to reply to you until now.
I do appreciate that you are making an effort to understand the U238 thought experiment. I regret to inform you that the U238 behavior is not the result of a unique process or bad use of the method, or measurement error. To understand what’s going on, you still need to understand probabilistic modeling. I think you can get there, but it’s going to take some work.
To help you along, I will mention that you need to understand the difference between a single event, on the one hand, and a large aggregation of events on the other. I will also mention that the decay of U238 can be described as a cumulative distribution function over time.
I am curious as to your reluctance to even attempt to answer the questions about lighting and the crime rate in Chicago neighborhoods. It is important for you to work on that problem, because it will help you understand another important aspect of probabilistic modeling. Why not give it a try, dcs?
The rest of your response demonstrates your fundamental misunderstanding of probabilistic modeling. Whenever I have described a theory whose predictions are based on probabilistic modeling, you have not been able to understand it. So let’s keep working on the U238 thought experiment and the Chicago crime rate data, shall we?
Peace,
I agree, dcs. Isn’t it preferable to ensure that you understand the methods with a less religiously-charged system before applying them to evolution?
ok chris. so i see that you continue to ignore my counter claims (i guess its because you cant falsified them, but its ok). i hope you understand now why your prediction isnt scientific.
peace.
Actually, my friend dcs, I have already answered them. Because they relied on an understanding of probabilistic modeling, though, you were unable to understand my answers. I’m trying to help you understand, but you don’t seem interested in learning. That’s OK, you should feel free to spend you time on what interests you. The world is a big place and God has given many different gifts to all of us. Since you are not interested in understanding science, by all means pursue something else and do it by God’s grace and power.
Grace and peace,
my friend chris- if you say so…
peace
Sorry to resurrect this thread, but I noticed that in @DennisVenema’s original article, there is very little discussion on whether or not Tomkins actually presented good evidence of the 150bp fragment of “vtg” being functional. It’s true that there’s a false dichotomy between “functional” and “pseudogene”, and I think it’s important to emphasise that point, but I think Tomkins’ claims of function should also be scrutinised.
Having read his short “paper”, I have to say that I’m astounded by how weak the evidence he presents is. There’s absolutely no “there” there.
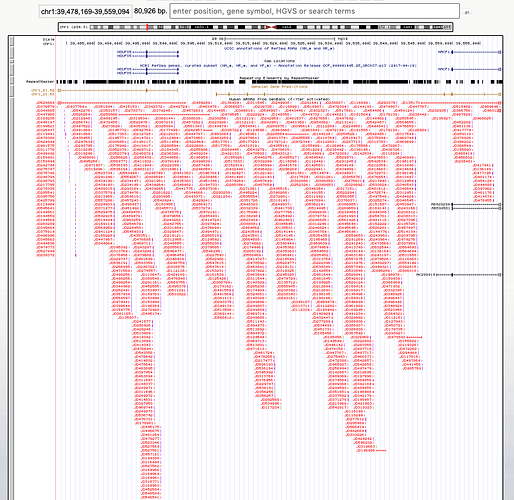
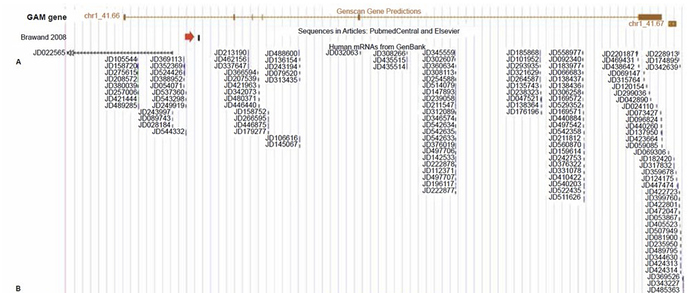
For example, his figure 2 presents gene expression data for what he claims is the “GAM gene”. He says that the very short transcripts dotted all over the place are indicative of some kind of active RNA gene, but if you search for the same set of transcripts from Bentwich (2007), Tomkins’ source for these supposedly functional “GAM” RNAs in other parts of the genome, you quickly realise that these transcripts show matches literally all over the genome. I picked a random location somewhere else on human chromosome 1 of approximately the same size as Tomkins’ “GAM gene”, and look what I found:
All those little red lines with labels are sequences that match the “GAM RNAs” identified by Bentwich (2007). Compare this to Tomkins’ figure (the “GAM RNAs” are in black rather than red):
Oh, and did I mention that Bentwich (2007) isn’t actually a published paper, but a patent? The patent application was made in 2002, and granted in 2007. Reference to these “GAM” genes online is practically non-existent aside from this patent and people discussing Tomkins article. No primary literature.
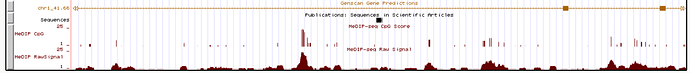
It’s a similar story for his claimed lowering of cytosine methylation specific to the supposed transcription factor binding site. What Tomkins shows:
What a zoomed-out view (15,000bp) looks like:
Note also that Tomkins only presents the raw MeDIP signal data because it visually supports his description at a small scale. He doesn’t show the MeDIP CpG score, which is basically the cleaned-up version of the same data, after removing noise etc. Suddenly the “lowering” around the vtg fragment disappears altogether, never mind at a particular “binding site”.
I could go on, but it’s late and I’ll give others a chance to jump in before I continue with the many, many flaws I can see with Tomkins’ “paper”.
Yes, this was clear to me then (and still is now) - but I decided to focus on the other aspects lest it be seen as an argument over the evidence. Even in the “worst case” scenario, he still doesn’t have an argument against common ancestry. That the “evidence” is so very weak only serves to show how willing he is to use poor arguments that support his views. If an evolutionary biologist offered up “evidence” of this caliber he would ridicule it.
Like I said, I think it’s important to make it clear to lay readers that Tomkins’ claims of functionality are utterly flawed, in addition to his dichotomy. As it stands, I don’t know of any good online rebuttals to Tomkins claims of function, which gives sites like AiG, ICR, and evolutionnews free reign to say things like (my emphases):
Tomkins has presented evidence that the VIT1 pseudogene sits in part of an intron that is transcribed and produces long non-coding RNAs of the type we know often have function. What does Venema say next? Does he contest this evidence for functionality? No. According to Venema the “major problem” with Tomkins’s argument isn’t the evidence for functionality in the VIT1 pseudogene.
Functional Pseuodogenes and Common Descent | Evolution News
Upon investigation, we see that this 150-base sequence is not an ancient egg-laying “fossil” in the human genome. It’s a functional enhancer element in a GAM gene expressed in brain tissues.
Evolutionists Lay an Egg: Vitellogenin Pseudogene Debunked | The Institute for Creation Research
Venema also takes exception to the latter half of Tomkins’ paper. In the latter half, Tomkins supplies evidence in favor of a function for the purported human vit1 gene fragment. Venema: “The major problem with this argument is that it subscribes to a false dichotomy: that this sequence is either a VIT1 pseudogene fragment or a functional part of another gene. From an evolutionary perspective, there is no issue with it being both.”18
Venema seems to have forgotten the challenge that he laid down a couple years prior: “I would invite these [creationist] groups, all of whom . . . suggest that ‘junk DNA’ is no longer a tenable idea, to ‘take the test’ and offer an explanation for the features we observe in the human Vitellogenin 1 pseudogene.”19 Tomkins has taken Venema’s test, just as Venema requested. Why is this element of Venema’s challenge suddenly no longer important? Venema seems to have moved the goalposts again.
Finding Adam in the Genome: A BioLogos cover-up? | Answers in Genesis
The average person investigating this subject will probably come away with the impression that Tomkins made this amazing new discovery that the human vitellogenin pseudogene is actually functional, and that you (and the rest of the “evolutionists”) have largely conceded this point.
I think the average reader of these sites believes that-
- “Evolutionary bias” is clouding the ability of scientists. Its no surprise a “compromising evolutionist” like Venema would miss this.
- With a separate creation of humans, all(?) of our genome was put there intentionally and has function. There is no ‘waste.’ There is no ‘junk.’
A side note: still don’t get why something having or not having function is relevant to common ancestry.
I don’t just mean the average AIG reader, I mean the average person investigating this subject, including non-creationists. That’s why I think it’s always important to get the information out there: for the fence-sitters, for the scientifically interested laypeople.
I think it’s a good idea to write on these topics. What do you mean by ‘fence-sitters?’ Surprisingly when I’ve done various google searches one of the first things I find are blog posts by EN (which in my opinion is purposefully deceptively named).
Fence-sitters are the ones who read these kinds of forums, watch debates, etc, and haven’t made their mind up about evolution. They don’t know what to believe yet, and can be impressionable.
WRT to EN coming top of your google search results, that’s probably a result of your personal search and browsing history. If you search for the same terms on a brand-new computer from a new IP address I don’t think they turn up very often.
I think there’s enough wrong with the paper to warrant me writing an extended blog post on the subject over on evograd.wordpress.com. I’m really busy at the moment, but maybe in a few weeks I’ll find time.
2 months later… I’m done!
Great article - very detailed. It covers a bunch of territory that I wanted to, back in the day, but lost patience for (and also wasn’t thought to be of a broad enough interest) - and also digs significantly deeper than I dug. Thanks for taking the time to go into this level of detail.
This topic was automatically closed 6 days after the last reply. New replies are no longer allowed.